Filtering signal in azimuthal space#
Usually, diffraction signal presents a polar symmetry, this means all pixel with the same azimuthal angle (χ) have similar intensities. The best way to exploit this is to take the mean, what is called azimuthal average. But the average is very sensitive to outlier, like gaps, missing pixels, shadows, cosmic rays or reflection coming from larger crystallite. In this tutorial we will see two alternative ways to remove those unwanted signal and focus on the majority of pixels: sigma clipping and median filtering.
[1]:
import os
import time
os.environ["PYOPENCL_COMPILER_OUTPUT"]="0"
import pyFAI
print(f"pyFAI version: {pyFAI.version}")
pyFAI version: 2025.12.0
[2]:
%matplotlib inline
from matplotlib.pyplot import subplots
from pyFAI.gui import jupyter
import numpy, fabio, pyFAI
from pyFAI import benchmark
from pyFAI.test.utilstest import UtilsTest
figsize = (10,5)
ai = pyFAI.load(UtilsTest.getimage("Pilatus6M.poni"))
img = fabio.open(UtilsTest.getimage("Pilatus6M.cbf")).data
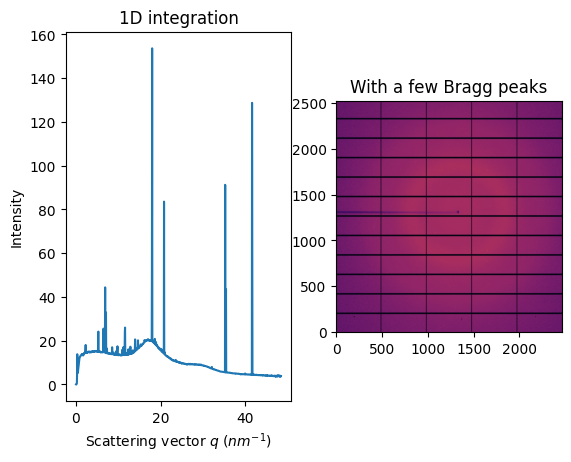
fig, ax = subplots(1, 2, figsize=figsize)
jupyter.display(img, ax=ax[1])
jupyter.plot1d(ai.integrate1d(img, 1000), ax=ax[0])
ax[1].set_title("With a few Bragg peaks");
WARNING:pyFAI.gui.matplotlib:Matplotlib already loaded with backend `inline`, setting its backend to `QtAgg` may not work!
Azimuthal sigma-clipping#
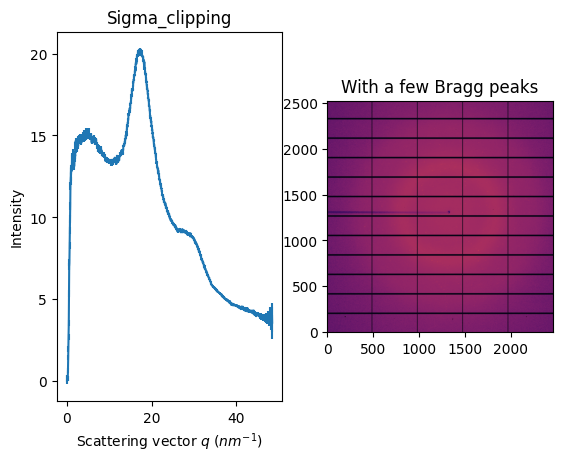
The idea is to discard pixels which look like outliers in the distribution of all pixels contributing to a single azimuthal bin. It requires an error model like poisson but it has been proven to be better to use the variance in the given azimuthal ring. All details are available in this publication: https://doi.org/10.1107/S1600576724011038 also available at https://doi.org/10.48550/arXiv.2411.09515
[3]:
fig, ax = subplots(1, 2, figsize=figsize)
jupyter.display(img, ax=ax[1])
jupyter.plot1d(ai.sigma_clip(img, 1000, error_model="hybrid", method=("no", "csr", "cython")), ax=ax[0])
ax[1].set_title("With a few Bragg peaks")
ax[0].set_title("Sigma_clipping");
Of course, sigma-clip takes several extra parameters like the number of iterations to perform, the cut-off, the error model, … There are alo a few limitation: * The algorithm needs to be the CSR-sparse matrix multiplication: since several integration are needed, it makes no sense to use an histogram based algorithm. * The algorithm is available with any implementation: Python (using scipy.saprse), Cython and OpenCL, and it runs just fine on GPU. * Sigma-clipping is incompatible with any kind of pixel splitting: With pixel splitting, a single pixel can contribute to several azimuthal bins and discarding a pixel in one ring could disable it in the neighboring ring (or not, since bins are processed in parallel).
Sigma-clipping performances:#
[4]:
method = ["no", "csr", "cython"]
[5]:
%%time
perfs_integrate_python = {}
perfs_integrate_cython = {}
perfs_integrate_opencl = {}
perfs_sigma_clip_python = {}
perfs_sigma_clip_cython = {}
perfs_sigma_clip_opencl = {}
for ds in pyFAI.benchmark.PONIS:
ai = pyFAI.load(UtilsTest.getimage(ds))
if ai.wavelength is None: ai.wavelength=1.54e-10
img = fabio.open(UtilsTest.getimage(pyFAI.benchmark.datasets[ds])).data
size = numpy.prod(ai.detector.shape)
print(ds)
print(" Cython")
meth = tuple(method)
nbin = max(ai.detector.shape)
print(" * integrate ", end="")
perfs_integrate_cython[size] = %timeit -o ai.integrate1d(img, nbin, method=meth)
print(" * sigma-clip", end="")
perfs_sigma_clip_cython[size] = %timeit -o ai.sigma_clip(img, nbin, method=meth, error_model="azimuthal")
print(" Python")
meth = tuple(method[:2]+["python"])
print(" * integrate ", end="")
perfs_integrate_python[size] = %timeit -o ai.integrate1d(img, nbin, method=meth)
print(" * sigma-clip", end="")
perfs_sigma_clip_python[size] = %timeit -o ai.sigma_clip(img, nbin, method=meth, error_model="azimuthal")
print(" OpenCL")
meth = tuple(method[:2]+["opencl"])
print(" * integrate ", end="")
perfs_integrate_opencl[size] = %timeit -o ai.integrate1d(img, nbin, method=meth)
print(" * sigma-clip", end="")
perfs_sigma_clip_opencl[size] = %timeit -o ai.sigma_clip(img, nbin, method=meth, error_model="azimuthal")
Pilatus1M.poni
Cython
* integrate 8.77 ms ± 1.44 ms per loop (mean ± std. dev. of 7 runs, 100 loops each)
* sigma-clip7.66 ms ± 495 μs per loop (mean ± std. dev. of 7 runs, 100 loops each)
Python
* integrate 9.87 ms ± 60.4 μs per loop (mean ± std. dev. of 7 runs, 100 loops each)
* sigma-clip158 ms ± 231 μs per loop (mean ± std. dev. of 7 runs, 10 loops each)
OpenCL
* integrate 662 μs ± 1.86 μs per loop (mean ± std. dev. of 7 runs, 1,000 loops each)
* sigma-clip2.36 ms ± 11.7 μs per loop (mean ± std. dev. of 7 runs, 100 loops each)
Pilatus2M.poni
Cython
* integrate 15.1 ms ± 1.33 ms per loop (mean ± std. dev. of 7 runs, 10 loops each)
* sigma-clip16.4 ms ± 1.64 ms per loop (mean ± std. dev. of 7 runs, 100 loops each)
Python
* integrate 34.7 ms ± 612 μs per loop (mean ± std. dev. of 7 runs, 10 loops each)
* sigma-clip552 ms ± 6.1 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
OpenCL
* integrate 1.08 ms ± 1.25 μs per loop (mean ± std. dev. of 7 runs, 1,000 loops each)
* sigma-clip5.78 ms ± 23.9 μs per loop (mean ± std. dev. of 7 runs, 100 loops each)
Eiger4M.poni
Cython
* integrate 25.9 ms ± 4.43 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
* sigma-clip31.9 ms ± 2.66 ms per loop (mean ± std. dev. of 7 runs, 10 loops each)
Python
* integrate 61.1 ms ± 211 μs per loop (mean ± std. dev. of 7 runs, 10 loops each)
* sigma-clip1.12 s ± 1.07 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
OpenCL
* integrate 1.83 ms ± 1.32 μs per loop (mean ± std. dev. of 7 runs, 1,000 loops each)
* sigma-clip10.4 ms ± 8.63 μs per loop (mean ± std. dev. of 7 runs, 100 loops each)
Pilatus6M.poni
Cython
* integrate 39.2 ms ± 4.8 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
* sigma-clip33.5 ms ± 1.9 ms per loop (mean ± std. dev. of 7 runs, 10 loops each)
Python
* integrate 82.4 ms ± 166 μs per loop (mean ± std. dev. of 7 runs, 10 loops each)
Compiler time: 0.22 s
* sigma-clip1.54 s ± 3.35 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
OpenCL
* integrate 2.39 ms ± 13 μs per loop (mean ± std. dev. of 7 runs, 100 loops each)
* sigma-clip13.4 ms ± 52.6 μs per loop (mean ± std. dev. of 7 runs, 100 loops each)
Eiger9M.poni
Cython
* integrate 61.1 ms ± 5.7 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
* sigma-clip64.1 ms ± 2.39 ms per loop (mean ± std. dev. of 7 runs, 10 loops each)
Compiler time: 0.14 s
Python
* integrate 150 ms ± 509 μs per loop (mean ± std. dev. of 7 runs, 10 loops each)
* sigma-clip2.87 s ± 6.63 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
OpenCL
* integrate 3.59 ms ± 12.1 μs per loop (mean ± std. dev. of 7 runs, 100 loops each)
* sigma-clip24.7 ms ± 27.1 μs per loop (mean ± std. dev. of 7 runs, 10 loops each)
Mar3450.poni
Cython
* integrate 57.8 ms ± 5.9 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
* sigma-clip62.5 ms ± 3.14 ms per loop (mean ± std. dev. of 7 runs, 10 loops each)
Python
* integrate 148 ms ± 482 μs per loop (mean ± std. dev. of 7 runs, 10 loops each)
* sigma-clip3.05 s ± 3.62 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
OpenCL
* integrate 4.03 ms ± 13.3 μs per loop (mean ± std. dev. of 7 runs, 100 loops each)
* sigma-clip27.5 ms ± 66.4 μs per loop (mean ± std. dev. of 7 runs, 10 loops each)
Fairchild.poni
Cython
* integrate 106 ms ± 2.84 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
* sigma-clip117 ms ± 3.66 ms per loop (mean ± std. dev. of 7 runs, 10 loops each)
Python
* integrate 376 ms ± 3.68 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
* sigma-clip9.16 s ± 76.7 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
OpenCL
* integrate 4 ms ± 7.14 μs per loop (mean ± std. dev. of 7 runs, 100 loops each)
* sigma-clip32.5 ms ± 148 μs per loop (mean ± std. dev. of 7 runs, 10 loops each)
CPU times: user 38min 46s, sys: 28.3 s, total: 39min 14s
Wall time: 5min 43s
[6]:
fig, ax = subplots()
ax.set_xlabel("Image size (Mpix)")
ax.set_ylabel("Frames per seconds")
sizes = numpy.array(list(perfs_integrate_python.keys()))/1e6
ax.plot(sizes, [1/i.best for i in perfs_integrate_python.values()], label="Integrate/Python", color='green', linestyle='dashed', marker='1')
ax.plot(sizes, [1/i.best for i in perfs_integrate_cython.values()], label="Integrate/Cython", color='orange', linestyle='dashed', marker='1')
ax.plot(sizes, [1/i.best for i in perfs_integrate_opencl.values()], label="Integrate/OpenCL", color='blue', linestyle='dashed', marker='1')
ax.plot(sizes, [1/i.best for i in perfs_sigma_clip_python.values()], label="Sigma-clip/Python", color='green', linestyle='dotted', marker='2')
ax.plot(sizes, [1/i.best for i in perfs_sigma_clip_cython.values()], label="Sigma-clip/Cython", color='orange', linestyle='dotted', marker='2')
ax.plot(sizes, [1/i.best for i in perfs_sigma_clip_opencl.values()], label="Sigma-clip/OpenCL", color='blue', linestyle='dotted', marker='2')
ax.set_yscale("log")
ax.legend()
ax.set_title("Performance of Sigma-clipping vs integrate");
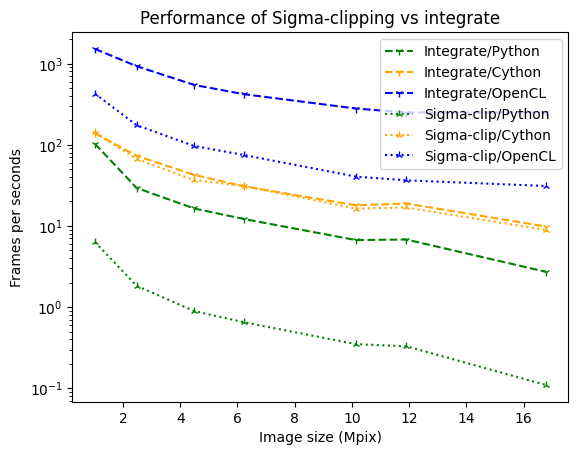
The penalties is very limited in Cython, much more in Python.
The biggest limitation of sigma-clipping is its incompatibility with pixel-splitting, feature needed when oversampling, i.e. taking many more points than the size of the diagonal of the image. While oversampling is not recommended in general case (due to the cross-corelation between bins it creates), it can be a nessessary evil, especially when performing Rietveld refinement where 5 points per peaks are needed, resolution that cannot be obtained with the pixel-size/distance couple accessible by the experimental setup.
Median filter in Azimuthal space#
The idea is to sort all pixels contibuting to an azimuthal bin and to average out all pixel between the lower and upper quantile. When those two thresholds are at one half, this filter provides actually the median. In order to be compatible with pixel splitting, each pixel is duplicated as many times as it contributes to different bins. After sorting fragments of pixels according to their normalization corrected signal, the cummulative sum of normalization is performed in order to detemine which fragments to average out.
[7]:
ai = pyFAI.load(UtilsTest.getimage("Pilatus6M.poni"))
img = fabio.open(UtilsTest.getimage("Pilatus6M.cbf")).data
method = ["full", "csr", "cython"]
percentile=(40,60)
pol=0.99
[8]:
fig, ax = subplots(1, 2, figsize=figsize)
jupyter.display(img, ax=ax[1])
jupyter.plot1d(ai.medfilt1d_ng(img, 1000, method=method, percentile=percentile, polarization_factor=pol), ax=ax[0])
ax[1].set_title("With a few Bragg peaks")
ax[0].set_title("Median filtering");
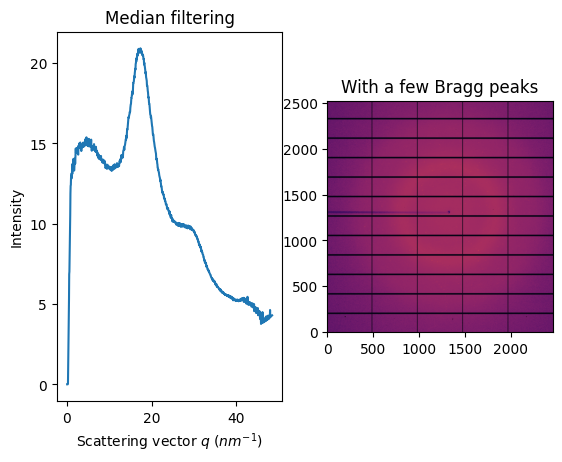
Unlike the sigma-clipping, this median filter does not equires any error model; but the computationnal cost induced by the sort is huge. In addition, the median is very sensitive and requires a good geometry and modelisation of the polarization.
[9]:
%%time
perf2_integrate_python = {}
perf2_integrate_cython = {}
perf2_integrate_opencl = {}
perf2_medfilt_python = {}
perf2_medfilt_cython = {}
perf2_medfilt_opencl = {}
for ds in pyFAI.benchmark.PONIS:
ai = pyFAI.load(UtilsTest.getimage(ds))
if ai.wavelength is None: ai.wavelength=1.54e-10
img = fabio.open(UtilsTest.getimage(pyFAI.benchmark.datasets[ds])).data
size = numpy.prod(ai.detector.shape)
print(ds)
print(" Cython")
meth = tuple(method)
nbin = max(ai.detector.shape)
print(" * integrate ", end="")
perf2_integrate_cython[size] = %timeit -o ai.integrate1d(img, nbin, method=meth)
print(" * medianfilter", end="")
perf2_medfilt_cython[size] = %timeit -o ai.medfilt1d_ng(img, nbin, method=meth)
print(" Python")
meth = tuple(method[:2]+["python"])
print(" * integrate ", end="")
perf2_integrate_python[size] = %timeit -o ai.integrate1d(img, nbin, method=meth)
print(" * medianfilter", end="")
perf2_medfilt_python[size] = %timeit -o ai.medfilt1d_ng(img, nbin, method=meth)
print(" OpenCL")
meth = tuple(method[:2]+["opencl"])
print(" * integrate ", end="")
perf2_integrate_opencl[size] = %timeit -o ai.integrate1d(img, nbin, method=meth)
print(" * medianfilter", end="")
perf2_medfilt_opencl[size] = %timeit -o ai.medfilt1d_ng(img, nbin, method=meth)
Pilatus1M.poni
Cython
* integrate 8.55 ms ± 1.52 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
* medianfilter24.5 ms ± 1.83 ms per loop (mean ± std. dev. of 7 runs, 10 loops each)
Python
* integrate 12.8 ms ± 129 μs per loop (mean ± std. dev. of 7 runs, 100 loops each)
* medianfilter1.26 s ± 19.5 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
OpenCL
* integrate 716 μs ± 3.01 μs per loop (mean ± std. dev. of 7 runs, 1,000 loops each)
* medianfilter9.37 ms ± 17.6 μs per loop (mean ± std. dev. of 7 runs, 100 loops each)
Pilatus2M.poni
Cython
* integrate 25.4 ms ± 4.35 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
* medianfilter68.7 ms ± 2.57 ms per loop (mean ± std. dev. of 7 runs, 10 loops each)
Python
* integrate 45.2 ms ± 557 μs per loop (mean ± std. dev. of 7 runs, 10 loops each)
* medianfilter4.34 s ± 57.9 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
OpenCL
* integrate 1.22 ms ± 3.43 μs per loop (mean ± std. dev. of 7 runs, 1,000 loops each)
* medianfilter30.5 ms ± 57.9 μs per loop (mean ± std. dev. of 7 runs, 10 loops each)
Eiger4M.poni
Cython
* integrate 37.1 ms ± 1.43 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
* medianfilter112 ms ± 2.11 ms per loop (mean ± std. dev. of 7 runs, 10 loops each)
Python
* integrate 83.8 ms ± 811 μs per loop (mean ± std. dev. of 7 runs, 10 loops each)
* medianfilter7.43 s ± 122 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
OpenCL
* integrate 1.99 ms ± 36.9 μs per loop (mean ± std. dev. of 7 runs, 100 loops each)
* medianfilter54 ms ± 21.9 μs per loop (mean ± std. dev. of 7 runs, 10 loops each)
Pilatus6M.poni
Cython
* integrate 36 ms ± 217 μs per loop (mean ± std. dev. of 7 runs, 1 loop each)
* medianfilter151 ms ± 3.67 ms per loop (mean ± std. dev. of 7 runs, 10 loops each)
Python
* integrate 112 ms ± 634 μs per loop (mean ± std. dev. of 7 runs, 10 loops each)
* medianfilter11.1 s ± 77.2 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
OpenCL
* integrate 2.66 ms ± 5.82 μs per loop (mean ± std. dev. of 7 runs, 100 loops each)
* medianfilter77.5 ms ± 67.6 μs per loop (mean ± std. dev. of 7 runs, 10 loops each)
Eiger9M.poni
Cython
* integrate 61.3 ms ± 6.3 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
* medianfilter233 ms ± 8.64 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
Python
* integrate 182 ms ± 2.22 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
* medianfilter18.3 s ± 103 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
OpenCL
* integrate 4.13 ms ± 32.1 μs per loop (mean ± std. dev. of 7 runs, 1 loop each)
* medianfilter138 ms ± 124 μs per loop (mean ± std. dev. of 7 runs, 10 loops each)
Mar3450.poni
Cython
* integrate 75.4 ms ± 774 μs per loop (mean ± std. dev. of 7 runs, 1 loop each)
* medianfilter258 ms ± 4.63 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
Python
* integrate 229 ms ± 4.55 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
* medianfilter21.1 s ± 66.5 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
OpenCL
* integrate 4.78 ms ± 21.8 μs per loop (mean ± std. dev. of 7 runs, 1 loop each)
* medianfilter161 ms ± 183 μs per loop (mean ± std. dev. of 7 runs, 10 loops each)
Fairchild.poni
Cython
* integrate 100 ms ± 4.07 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
* medianfilter384 ms ± 4.64 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
Python
* integrate 305 ms ± 2.22 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
* medianfilter25.8 s ± 181 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
OpenCL
* integrate 4.61 ms ± 61.2 μs per loop (mean ± std. dev. of 7 runs, 1 loop each)
* medianfilter172 ms ± 178 μs per loop (mean ± std. dev. of 7 runs, 10 loops each)
CPU times: user 25min 21s, sys: 26 s, total: 25min 47s
Wall time: 14min 42s
[10]:
fig, ax = subplots()
ax.set_xlabel("Image size (Mpix)")
ax.set_ylabel("Frames per seconds")
sizes = numpy.array(list(perf2_integrate_python.keys()))/1e6
ax.plot(sizes, [1/i.best for i in perf2_integrate_python.values()], label="Integrate/Python", color='green', linestyle='dashed', marker='1')
ax.plot(sizes, [1/i.best for i in perf2_integrate_cython.values()], label="Integrate/Cython", color='orange', linestyle='dashed', marker='1')
ax.plot(sizes, [1/i.best for i in perf2_integrate_opencl.values()], label="Integrate/OpenCL", color='blue', linestyle='dashed', marker='1')
ax.plot(sizes, [1/i.best for i in perf2_medfilt_python.values()], label="Medfilt/Python", color='green', linestyle='dotted', marker='2')
ax.plot(sizes, [1/i.best for i in perf2_medfilt_cython.values()], label="Medfilt/Cython", color='orange', linestyle='dotted', marker='2')
ax.plot(sizes, [1/i.best for i in perf2_medfilt_opencl.values()], label="Medfilt/OpenCL", color='blue', linestyle='dotted', marker='2')
ax.set_yscale("log")
ax.legend()
ax.set_title("Performance of Median filtering vs integrate");
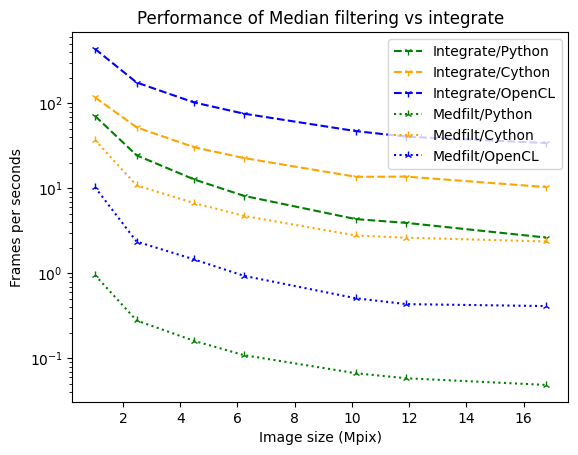
As one can see, the penalities are much larger for OpenCL and Python than for Cython.
Conclusion#
Sigma-clipping and median-filtering are alternatives to azimuthal integration and offer the ability to reject outliers. They are not more difficult to use but slightly slower owing to their greater complexity.