ImXPAD S540 detector at D2AM#
This tutorial corresponds to the calibration the goniometer an ImXPAD detector composed of 8 stripes of 7 modules, many of which are defective, on a goniometer.
This detector is mounted on the goniometer 2theta arm at the D2AM beam-line, French CRG at the ESRF synchrotron.
The raw data files are available at: http://www.silx.org/pub/pyFAI/gonio/D2AM-15/
[1]:
%matplotlib inline
#For documentation purpose, `inline` is used to enforce the storage of images into the notebook
# %matplotlib widget
import numpy
from matplotlib.pyplot import subplots
[2]:
import os, time
start_time = time.perf_counter()
import fabio, pyFAI
print(f"Working with pyFAI version {pyFAI.version}")
from pyFAI.goniometer import GeometryTransformation, GoniometerRefinement, Goniometer
from pyFAI.gui import jupyter
from math import ceil
Working with pyFAI version 2025.12.0-beta0
[3]:
#Download all data
from silx.resources import ExternalResources
#Comment out and configure the proxy if you are behind a firewall
#os.environ["http_proxy"] = "http://proxy.company.com:3128"
downloader = ExternalResources("pyFAI", "http://www.silx.org/pub/pyFAI/testimages", "PYFAI_DATA")
all_files = downloader.getdir("LaB6_gonio_D2AM.tar.bz2")
print("List of files downloaded:")
for i in all_files:
print(" "+os.path.basename(i))
detector_file = [i for i in all_files if i.endswith("D5Geom-2018.h5")][0]
images = [i for i in all_files if i.endswith(".edf")]
npt_files = [i for i in all_files if i.endswith(".npt")]
List of files downloaded:
LaB6_gonio_D2AM
16Dec08D5_1777-rsz.npt
16Dec08D5_1791-rsz.npt
16Dec08D5_1729-rsz.edf
16Dec08D5_1763-rsz.npt
D5Geom-2018.h5
16Dec08D5_1725-rsz.edf
16Dec08D5_1728-rsz.npt
16Dec08D5_1784-rsz.npt
16Dec08D5_1763-rsz.edf
16Dec08D5_1728-rsz.edf
16Dec08D5_1735-rsz.npt
16Dec08D5_1727-rsz.npt
16Dec08D5_1729-rsz.npt
16Dec08D5_1770-rsz.edf
16Dec08D5_1784-rsz.edf
16Dec08D5_1730-rsz.npt
16Dec08D5_1742-rsz.npt
16Dec08D5_1770-rsz.npt
16Dec08D5_1725-rsz.npt
16Dec08D5_1726-rsz.npt
16Dec08D5_1777-rsz.edf
16Dec08D5_1756-rsz.edf
16Dec08D5_1735-rsz.edf
16Dec08D5_1730-rsz.edf
16Dec08D5_1742-rsz.edf
16Dec08D5_1791-rsz.edf
16Dec08D5_1749-rsz.npt
16Dec08D5_1726-rsz.edf
16Dec08D5_1756-rsz.npt
16Dec08D5_1727-rsz.edf
16Dec08D5_1749-rsz.edf
[4]:
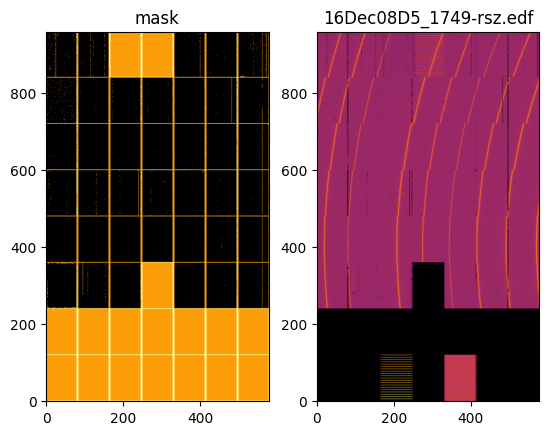
#Definition of the detector and deplay of an image and its mask:
d5 = pyFAI.detector_factory(detector_file)
print("Detector shape: ",d5.shape)
fimg = fabio.open(images[-1])
for k,v in fimg.header.items():
print(k, ": ", v)
f, ax = subplots(1, 2)
jupyter.display(d5.mask, label="mask", ax=ax[0])
jupyter.display(fimg.data, label=os.path.basename(fimg.filename), ax=ax[1]);
Detector shape: (960, 578)
EDF_DataBlockID : 0.Image.Psd
EDF_BinarySize : 4439040
EDF_HeaderSize : 1536
ByteOrder : LowByteFirst
DataType : DoubleValue
Dim_1 : 578
Dim_2 : 960
Image : 0
HeaderID : EH:000000:000000:000000
Size : 4439040
VersionNumber : 1
Epoch : 1481327234.3919599056
det_sample_dist : 0
y_beam : 0
x_beam : 0
Lambda : 0.495938
offset : 0
count_time : 120
point_no : 24
scan_no : 906
preset : 0
col_end : 559
col_beg : 0
row_end : 959
row_beg : 0
counter_pos : 120 2689 156 15.3097 0 90.3692 15.3097 0 25 25 1749 5.65726e+09 0 0 0 174.925 0 0 15.3097
counter_mne : sec vct1 vct2 vct3 vct4 Imach pseudoC pfoil Emono Ecod img roi1 roi2 roi3 roi4 pico1 pico2 pico3 pico4
motor_pos : 24.0001 0.077502 89.9912 -89.9921 -0.0032 0.0022 57.1197 134.748 -32.9504 0.16656 -5 0.47558 -1.5 0 0 0 4.53604 0.1416 1.04 1.04022 1.04022 -4.4 -1.10211 -0.543725 -9.962 -14.038 -16.865 -7.195 24 -2.038 24.06 4.835 1
motor_mne : del eta chi phi nu mu keta kap kphi tsx tsy tsz rox roy tox toy mono inc1 courb courbb courbf omega khimono gamma su6 sd6 sf6 sb6 vg6 vo6 hg6 ho6 rien
suffix : .edf
prefix : 16Dec08D5_
dir : /users/opd02/raw
run : 1749
title : CCD Image
[5]:
# Define wavelength and create our "large" LaB6 calibrant
wavelength = 0.495938 * 1e-10
from pyFAI.calibrant import get_calibrant
LaB6 = get_calibrant("LaB6")
LaB6.wavelength = wavelength
print("2theta max: ", numpy.degrees(LaB6.get_2th()[-1]))
print("Number of reflections: ", len(LaB6.get_2th()))
2theta max: 179.17349767223402
Number of reflections: 236
[6]:
#Use a few manually calibrated images:
npt_files.sort()
print("Number of hand-calibrated images :",len(npt_files))
Number of hand-calibrated images : 15
[7]:
# Definition of the goniometer translation function:
# The detector rotates vertically, around the horizontal axis, i.e. rot2.
# Rotation both around axis 1 and axis 2 are allowed
goniotrans = GeometryTransformation(param_names = ["dist", "poni1", "poni2",
"rot1", "rot2", "rot3", "scale1", "scale2" ],
dist_expr="dist",
poni1_expr="poni1",
poni2_expr="poni2",
rot1_expr="scale1 * pos +rot1",
rot2_expr="scale2 * pos + rot2",
rot3_expr="rot3")
#Definition of the function reading the goniometer angle from the filename of the image.
def get_angle(metadata):
"""Takes the angle from the first motor position and returns the angle of the goniometer arm"""
return float(metadata["motor_pos"].split()[0])
print('filename', os.path.basename(fimg.filename), "angle:",get_angle(fimg.header))
filename 16Dec08D5_1749-rsz.edf angle: 24.0001
[8]:
# Definition of the geometry refinement: the parameter order is the same as the param_names
rot3 = numpy.pi/2
scale1 = -numpy.pi/180
scale2 = 0
param = {"dist":0.5,
"poni1":0.05,
"poni2":0.05,
"rot1":0,
"rot2":0,
"rot3": rot3,
"scale1": scale1,
"scale2": scale2,
}
#Defines the bounds for some variables
bounds = {"dist": (0.2, 0.8),
"poni1": (0, 0.1),
"poni2": (0, 0.1),
"rot1": (-0.1, 0.1),
"rot2": (-0.1, 0.1),
"rot3": (rot3, rot3), #strict bounds on rot3
"scale1": (scale1, scale1),
"scale2": (scale2, scale2),
}
gonioref = GoniometerRefinement(param, #initial guess
bounds=bounds,
pos_function=get_angle,
trans_function=goniotrans,
detector=d5, wavelength=wavelength)
print("Empty refinement object:", gonioref)
#Let's populate the goniometer refinement object with all control point files:
for fn in npt_files[:]:
base = os.path.splitext(fn)[0]
fimg = fabio.open(base + ".edf")
basename = os.path.basename(base)
sg =gonioref.new_geometry(basename, image=fimg.data, metadata=fimg.header, control_points=fn, calibrant=LaB6)
print(basename, "Angle:", sg.get_position())
print("Filled refinement object:")
print(gonioref)
Empty refinement object: GoniometerRefinement with 0 geometries labeled: .
16Dec08D5_1725-rsz Angle: -0.003
16Dec08D5_1726-rsz Angle: 0.9998
16Dec08D5_1727-rsz Angle: 2.0
16Dec08D5_1728-rsz Angle: 2.9998
16Dec08D5_1729-rsz Angle: 4.0002
16Dec08D5_1730-rsz Angle: 4.9998
16Dec08D5_1735-rsz Angle: 10.0001
16Dec08D5_1742-rsz Angle: 16.9996
16Dec08D5_1749-rsz Angle: 24.0001
16Dec08D5_1756-rsz Angle: 30.9997
16Dec08D5_1763-rsz Angle: 37.9999
16Dec08D5_1770-rsz Angle: 44.9997
16Dec08D5_1777-rsz Angle: 52.0
16Dec08D5_1784-rsz Angle: 58.9995
16Dec08D5_1791-rsz Angle: 65.9999
Filled refinement object:
GoniometerRefinement with 15 geometries labeled: 16Dec08D5_1725-rsz, 16Dec08D5_1726-rsz, 16Dec08D5_1727-rsz, 16Dec08D5_1728-rsz, 16Dec08D5_1729-rsz, 16Dec08D5_1730-rsz, 16Dec08D5_1735-rsz, 16Dec08D5_1742-rsz, 16Dec08D5_1749-rsz, 16Dec08D5_1756-rsz, 16Dec08D5_1763-rsz, 16Dec08D5_1770-rsz, 16Dec08D5_1777-rsz, 16Dec08D5_1784-rsz, 16Dec08D5_1791-rsz.
[9]:
# Initial refinement of the goniometer model with 5 dof
gonioref.refine3()
Free parameters: ['dist', 'poni1', 'poni2', 'rot1', 'rot2']
Fixed: {'rot3': 1.5707963267948966, 'scale1': -0.017453292519943295, 'scale2': 0}
message: Optimization terminated successfully
success: True
status: 0
fun: 3.46909185579596e-07
x: [ 5.225e-01 8.731e-02 4.574e-02 3.270e-03 -3.865e-02]
nit: 25
jac: [ 2.233e-08 4.963e-08 -2.134e-07 -1.039e-07 2.643e-08]
nfev: 151
njev: 25
multipliers: []
Constrained Least square 0.0003076612160235168 --> 3.46909185579596e-07
maxdelta on rot2: 0.0 --> -0.03864790947424475
[9]:
np.float64(3.46909185579596e-07)
[10]:
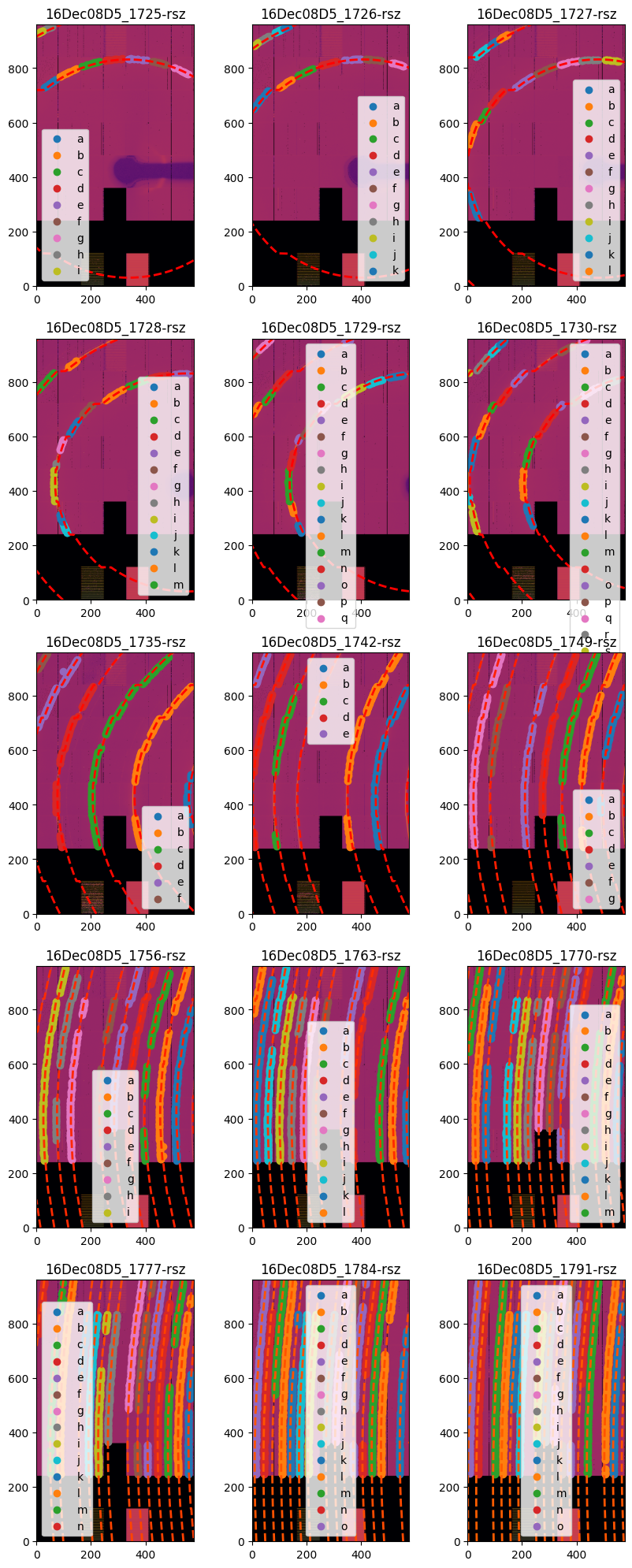
width = 3
height = int(ceil(len(gonioref.single_geometries) / width))
fig,ax = subplots(height, width,figsize=(10, 25))
for idx, sg in enumerate(gonioref.single_geometries.values()):
sg.geometry_refinement.set_param(gonioref.get_ai(sg.get_position()).param)
jupyter.display(sg=sg, ax=ax[idx//width, idx%width]);
[11]:
# Final pass of refinement with all constrains removed, very fine refinement
gonioref.bounds = None
gonioref.refine3(fix=["rot3"], method="slsqp", eps=1e-13, maxiter=10000, ftol=1e-12)
gonioref.refine3(fix=["rot3"], method="simplex", maxiter=10000, ftol=1e-12)
Free parameters: ['dist', 'poni1', 'poni2', 'rot1', 'rot2', 'scale1', 'scale2']
Fixed: {'rot3': 1.5707963267948966}
WARNING:pyFAI.goniometer:No bounds for optimization method Nelder-Mead
message: Optimization terminated successfully
success: True
status: 0
fun: 1.8635169200615737e-07
x: [ 5.217e-01 8.731e-02 4.574e-02 4.502e-03 -3.883e-02
-1.748e-02 -4.289e-05]
nit: 12
jac: [ 5.001e-08 0.000e+00 0.000e+00 -1.668e-08 1.720e-08
-1.627e-06 -7.544e-08]
nfev: 102
njev: 12
multipliers: []
Constrained Least square 3.46909185579596e-07 --> 1.8635169200615737e-07
maxdelta on rot1: 0.0032695604402495767 --> 0.004501663994410485
Free parameters: ['dist', 'poni1', 'poni2', 'rot1', 'rot2', 'scale1', 'scale2']
Fixed: {'rot3': 1.5707963267948966}
/users/kieffer/.venv/py314/lib/python3.14/site-packages/pyFAI/goniometer.py:1001: OptimizeWarning: Unknown solver options: ftol
res = minimize(self.residu3, param, method=method,
message: Optimization terminated successfully.
success: True
status: 0
fun: 1.389369732301749e-08
x: [ 5.202e-01 5.791e-02 4.048e-02 -4.767e-03 1.830e-02
-1.747e-02 -4.651e-04]
nit: 1027
nfev: 1747
final_simplex: (array([[ 5.202e-01, 5.791e-02, ..., -1.747e-02,
-4.651e-04],
[ 5.202e-01, 5.791e-02, ..., -1.747e-02,
-4.651e-04],
...,
[ 5.202e-01, 5.791e-02, ..., -1.747e-02,
-4.651e-04],
[ 5.202e-01, 5.791e-02, ..., -1.747e-02,
-4.651e-04]], shape=(8, 7)), array([ 1.389e-08, 1.389e-08, 1.389e-08, 1.389e-08,
1.389e-08, 1.389e-08, 1.389e-08, 1.389e-08]))
Constrained Least square 1.8635169200615737e-07 --> 1.389369732301749e-08
maxdelta on rot2: -0.038825381776677576 --> 0.01830309774699841
[11]:
np.float64(1.389369732301749e-08)
[12]:
#Create a MultiGeometry integrator from the refined geometry:
angles = []
images = []
for sg in gonioref.single_geometries.values():
angles.append(sg.get_position())
images.append(sg.image)
multigeo = gonioref.get_mg(angles)
multigeo.radial_range=(0, 80)
print(multigeo)
MultiGeometry integrator with 15 geometries on (0, 80) radial range ((2th_deg, chi_deg)) and (-180, 180) azimuthal range (deg)
[13]:
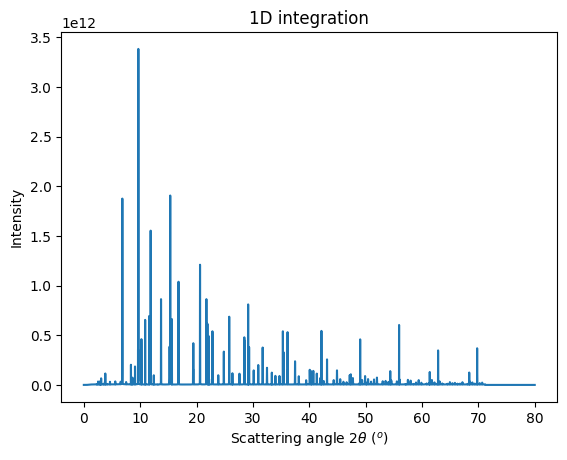
# Integrate the whole set of images in a single run:
res = multigeo.integrate1d(images, 10000)
jupyter.plot1d(res)
#Note the large number of peaks due to hot pixels ....
[13]:
<Axes: title={'center': '1D integration'}, xlabel='Scattering angle $2\\theta$ ($^{o}$)', ylabel='Intensity'>
[14]:
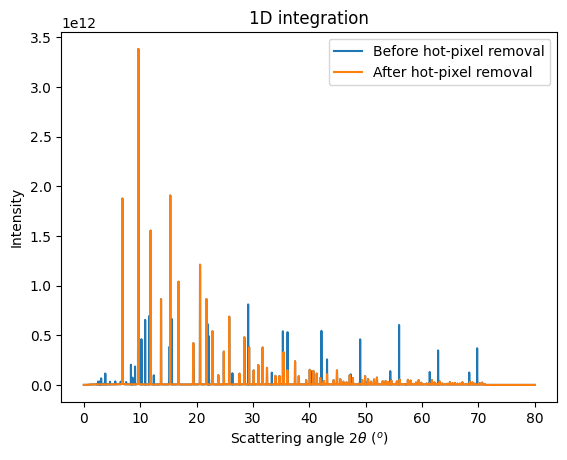
#Add hot pixels to the mask: pixel which are 15x more intense than the median in their ring.
thres = 15
old_mask = d5.mask.astype("bool", copy=True)
new_mask = d5.mask.astype("bool", copy=True)
for ai,img in zip(multigeo.ais,images):
b,a = ai.separate(img, 1000, restore_mask=0)
b[old_mask] = 0
b[b<0] = 0
# print(sum(b>thres*a))
new_mask = numpy.logical_or(new_mask, (b>thres*a))
print(#" Size of old mask", sum(old_mask),
#" Size of new mask",sum(new_mask),
" Number of pixel discarded", sum(new_mask)-sum(old_mask))
Number of pixel discarded [0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 2 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0]
[15]:
# Update the mask
for ai in multigeo.ais:
ai.detector.mask = new_mask
# Integrate the whole set of images in a single run:
res2 = multigeo.integrate1d(images, 10000)
ax = jupyter.plot1d(res, label="Before hot-pixel removal")
ax.plot(*res2, label="After hot-pixel removal")
ax.legend();
[16]:
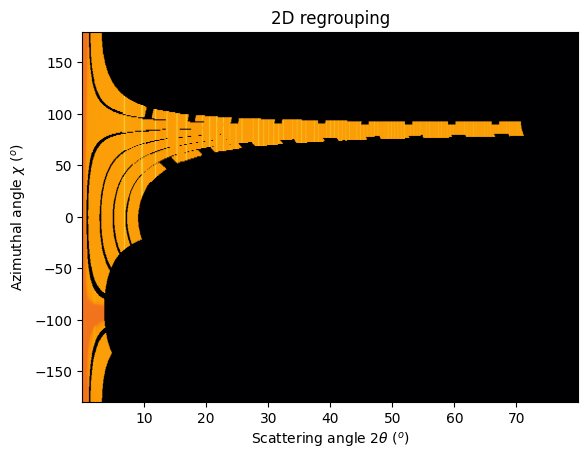
# Integrate the whole set of images in 2D:
res2d = multigeo.integrate2d(images, 1000, 360)
jupyter.plot2d(res2d);
[17]:
print(f"Total execution time {time.perf_counter()-start_time:.3f} s")
Total execution time 295.255 s